Spatialomics and AI:
A new age for personalised clinical medicine
Advances in spatialomics, including spatial transcriptomics, proteomics, and epigenomics, have provided researchers and clinicians with an unprecedented ability to glimpse into biological processes occurring in single cells and tissue microdomains.
Parallel advances in computation and artificial intelligence (AI) have enabled analyses of the vast amounts of data spatialomics produces, as well as testing of complex hypotheses requiring synthesis and interpretation of high-dimensional single- and multi-modal spatial data sets (multi-omics).
Together, spatialomics and AI are impacting the practice of medicine and have given rise to a new paradigm of science coined ‘spatial medicine’, where spatial biology insights play key roles in drug discovery and clinical decision-making.
Spatialomics and AI are making medicine more personal
A key area where spatialomics and AI are contributing to the transformation of medicine is in the transition from medicine guided by population-based estimates of risk to personalised medicine.
Traditionally, medical care has been guided using a population-based approach, which considers the clinical risk factors of groups of individuals who are most prone to developing a medical condition or who are already afflicted with a medical condition. Therapeutics are developed, interventional procedures and preventative screening intervals are optimised, and laboratory tests are designed to improve health outcomes for that group of individuals based on what is known about the group as a whole.
That care is typically used as the standard for an entire population of individuals without clarity as to whether it meets risk-based needs for specific individuals or whether their health outcomes will be improved by the recommended therapeutics, interventions, and tests.
Personalised medicine offers a potential solution to the insufficiencies of population-based medicine. Instead of using population-level estimates, it is designed to consider the unique biology and risk factors of each individual patient to help inform clinical decisions and ensure that care is delivered to each patient in a personalised manner.
Examples of personalised medicine range from adjunct tests, which analyse the expression of genes or other biomarkers in patient tissues to guide treatment decisions, to the development of personalised immune cell therapies based on mutations found in an individual patient’s tumour.
As personalised medicine continues to evolve, emerging technologies like spatialomics and AI are paving the way for laboratories to develop the next generation of clinical tests focused on individualised care.
Many studies have identified diagnostic biomarkers and signatures of disease progression and responses to therapies using spatialomics-based assays. However, few assays have been translated into clinical tests that can be adopted for routine clinical use. One area where a spatialomics test that utilises AI has been translated into the clinic to provide personalised medicine is in the management of Barrett’s oesophagus (BE).
Patients with Barrett’s oesophagus can benefit from personalised medicine
BE is the sole precursor to oesophageal adenocarcinoma (EAC). It develops in response to chronic gastrointestinal reflux disease (GERD) and results in metaplastic changes to the lining of the oesophagus.
Most patients with BE will not progress to EAC; however, those that do progress have a dismal five-year overall survival rate of only 22%. Historically, risk factors for BE have included: male sex, Caucasian race, over 50 years of age, obesity, a history of tobacco smoking, a family history of BE or EAC, and a history of GERD symptoms. Clinical guidelines generally recommend that individuals with combinations of these risk factors be screened for BE through endoscopies and collection of pinch biopsies.
Biopsy samples obtained during endoscopy are stained with haematoxylin and eosin (H&E) and assessed by a pathologist who reports whether a patient has normal oesophageal squamous epithelium, non-dysplastic (ND) BE, low-grade dysplasia (LGD), high-grade dysplasia (HGD), or EAC.
If the pathologist cannot confidently determine the grade of dysplasia for BE, they will typically report indefinite for dysplasia (IND). Traditionally, pathologists’ diagnostic findings have been used to estimate the patient’s risk of progressing to EAC and to establish follow-up preventative surveillance intervals or interventions.
This population-based estimate of risk is not appropriate for all patients, especially women, non-Caucasian, younger, and non-obese individuals without a history of GERD who can also be at high risk of developing BE and EAC, and it does not provide a patient’s individualised risk of progressing to EAC. The use of such a population-based approach makes determining which patients are at risk of progressing to EAC, and which are not, a significant challenge for clinicians.
Delivering personalised care to patients with Barrett’s oesophagus
To address this critical challenge facing clinicians, tools that incorporate personalised risk stratification into clinical practice are needed. For example, TissueCypher – a clinically available prognostic test – was designed to address this challenge.
The test combines spatialomics and AI to objectively determine each patient’s unique risk of their BE progressing to HGD or EAC within five years. It begins with a patient’s biopsied BE tissue undergoing multiplexed immunofluorescent labelling of nine protein biomarkers associated with key pathways in neoplastic progression to EAC, including tumour suppression, cell cycle control, angiogenesis, and immune cell infiltration.
Slides are then digitised by whole slide imaging and automatically processed by proprietary image analysis software that includes densely connected neural networks that identify BE tissue while excluding tissue processing artefacts and non-BE tissues. Additional computer vision algorithms identify and segment seven key cell and tissue structures to quantify 15 spatialomic features, which are not readily observable by the human eye.
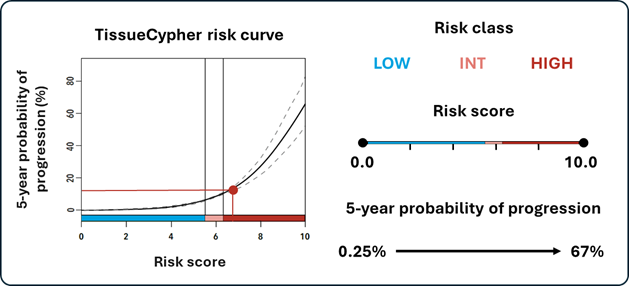
These feature values are then integrated into a locked risk stratification algorithm to produce the patient’s personalised risk report, including a risk score (0-10), a risk class (low, intermediate, high), and their probability of progression to HGD/EAC within five years.
Image: TissueCypher provides a risk report containing a risk curve, as well as a patient’s risk class, risk score, and five-year probability of progression to HGD/EAC.
TissueCypher’s personalised approach to determining each patient’s unique risk of progressing to HGD/EAC has been extensively validated through multiple independent and international studies.
Informing clinical decisions utilising data from AI-driven, spatialomic tests, such as TissueCypher, illustrates the advantages of personalised medicine.
Spatialomics and AI will continue to shape the future of medicine
The use of spatialomics and AI in clinical tests is contributing to a paradigm shift where personalised medicine is replacing population-based medicine across the medical landscape. With ongoing innovation in spatial biology and AI, spatialomics tests are poised for greater clinical availability.
Data from such tests, when combined with other biologic, spatialomic, and clinical datasets, may serve as the foundation for predictive models of disease at the cell, tissue, and organism level. These models could ultimately enable the creation of digital ‘twins’ of patients for their medical conditions, which can be mined for diagnostic, predictive, and prognostic insights to inform personalised clinical management plans.
We believe the future of medicine is personalised and that spatialomics and AI are helping to bridge the gap to create such a future.
About the authors
Dr Grant Daskivich serves as a medical writer (R&D, GI) at Castle Biosciences. Dr Daskivich joined Castle Biosciences in 2025 after working as a postdoctoral researcher at the University of Pittsburgh School of Medicine studying alpha-synuclein’s physiological role at the neuronal synapse. He earned his PhD in Molecular and Cell Biology at the University of Pittsburgh studying protein quality control and protein degradation in the endoplasmic reticulum. Dr Daskivich is an author of multiple scientific publications and has received awards for excellence in undergraduate teaching and mentorship.
Dr Erik Martin serves as director of spatialomics & gastrointestinal (gi) research & development (R&D) at Castle Biosciences. Dr Martin joined Castle Biosciences in 2024 after serving as a lead scientist at Booz Allen Hamilton. He completed postdoctoral training at the National Institute on Aging, a division of the US National Institutes of Health, where his spatialomics studies revealed novel insights into NF-kappaB pathway activity in single cells. Dr Martin earned his PhD in Molecular Medicine at the University of Maryland School of Medicine. He is also an inventor on several patents and the author of multiple scientific publications.
Supercharge your pharma insights: Sign up to pharmaphorum's newsletter for daily updates, weekly roundups, and in-depth analysis across all industry sectors.
Want to go deeper?
Continue your journey with these related reads from across pharmaphorum
Click on either of the images below for more articles from this edition of Deep Dive: AI 2025