Data interpretation of NGS data in biomedical context

Mark Hughes
Genomatix
In our personalized medicine focus month, Mark Hughes discusses data interpretation of next generation sequencing in the context of the biomedical industry.
The advance of Next Generation Sequencing (NGS) technology has enabled researchers to generate genome wide sequencing data of unprecedented quality and quantity. Chromatin IP (ChIP-Seq), Genotyping (DNA-Seq), expression analysis (RNA-Seq), microRNA expression (mRNA-Seq) and more recently Chromosome Confirmation Capture (3C, 4C and 5C) are the main application areas currently used for research purposes and also moving into the clinical setting.
 ,
"...the need for efficient analyses, data reduction and comprehensible visualizations has become critical..."
 ,
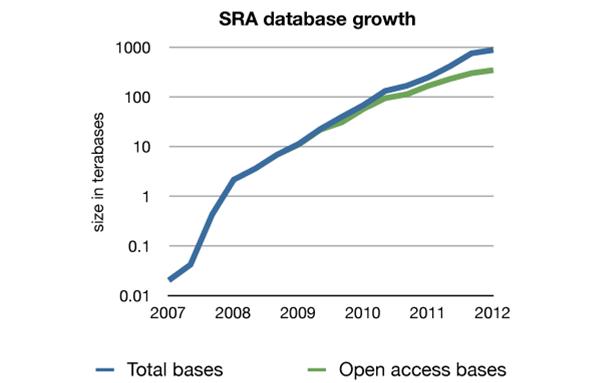
These applications are used in various research fields like epigenomics, transcriptomics and the analysis of structural variants. With data generation growing at an exponential rate (figure 1), the need for efficient analyses, data reduction and comprehensible visualizations has become critical, as well as the biomedical interpretation of NGS data.
Figure 1: The increase in contents of the Sequence Read Archive (SRA) (based on the numbers available at the overview page of the NCBI http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi as of 09/10/2012)
Turning data into meaning
After getting the raw files from an NGS sequencer, in either sequence or colour space files, further analysis generally starts with the alignment of reads to a reference genome. This is either whole genome or specially constructed references, e.g. transcriptome, splice junction, or targeted region.
The subsequent analysis of the dataset is to compare them to reference or a control sample (e.g. SNPs, re-arrangements, or differential expression of RNA or mRNA). Following this are then application-specific steps, an example being a copy number analysis, transcript analysis or the identification of areas of higher and / or lower read density as in ChIP-Seq.
 ,
"Turning the data into meaningful results, however, requires an interpretive step to layer the biological meaning around the data..."
 ,
Yet, these and other first steps are simply data conversion and reduction that shrinks the massive amount of raw data to a (usually still huge) number of regions or positions that may still contain a large amount of data that does not relate to the biological question in hand. While these steps are demanding in terms of processing power and storage requirements, they can be automated and pipelined for high-throughput analyses.
Turning the data into meaningful results, however, requires interpretive downstream analysis steps to layer the biological meaning around the data, and to achieve this a significant amount of high quality background information at several different levels is required (figure 2).
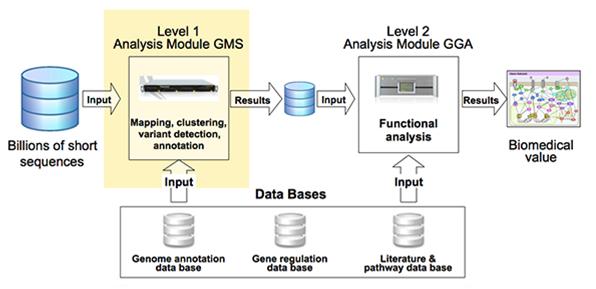 Figure 2: Pipeline for data analysis for biomedical results
Figure 2: Pipeline for data analysis for biomedical results
For example, to annotate regions of relevance, a comprehensive genome annotation database must be available to allow correlations with multiple types of genomic elements such as transcripts, transcription start sites and / or regulatory regions (promoters / enhancers) (figure 3).
 Figure 3: Screenshot from Genomatix Genome Browser showing incorporation of annotation with data obtained from NGS experiments.
Figure 3: Screenshot from Genomatix Genome Browser showing incorporation of annotation with data obtained from NGS experiments.
Combining RNA and genomic level, one can get detailed insight in molecular processes, e.g. proteins encoded by a specific transcript variant or even by structural variants of the genome caused by diseases like cancer (figure 4). This again requires a comprehensive database of available transcripts and splice junctions plus methods to detect structural variants and a visualization to make the intricate correlations between these data understandable.
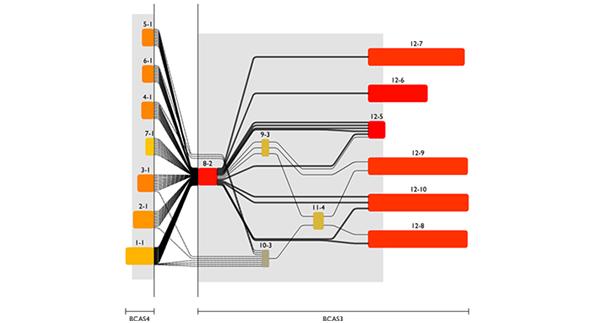 Fig 4: Screenshot from the Genomatix Transcriptome Viewer showing transcript fusions between the BCAS4 and BCAS3 genes in a breast cancer cell line.
Fig 4: Screenshot from the Genomatix Transcriptome Viewer showing transcript fusions between the BCAS4 and BCAS3 genes in a breast cancer cell line.
Understanding regulatory events triggered by the binding of transcription factor proteins requires yet another different level of database information, in this case transcription factor binding motifs and suitable algorithms to find binding site patterns within the sequence data (figure 5).
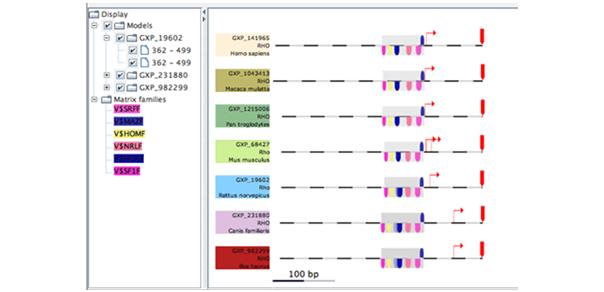 Fig 5: Screenshot showing a framework of transcription factors conserved between different species.
Fig 5: Screenshot showing a framework of transcription factors conserved between different species.
As these above mentioned elements are all part of a system and rarely act on their own, the integration into a network or pathway system is a reasonable subsequent move. In addition to connections between genes, categories like diseases, tissues, molecular processes, etc. can be used for association and filtering. Moreover, any gene-based information from literature or external databases can be incorporated, so researchers can easily move between an overview of the whole network to detailed information on gene level (figure 6).
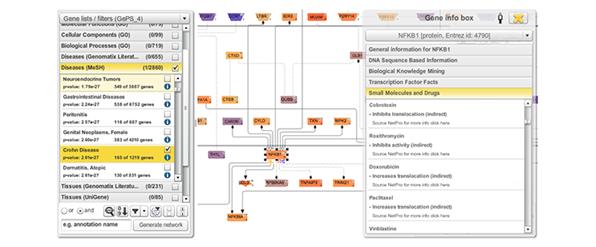 Figure 6: Excerpt from a network of genes associated with Crohn Disease. On the right hand side a list of small molecules and drugs can be seen that is associated with the transcription factor NFKB1. Different colourings of genes refer to their expression level derived from an RNA-Seq experiment.
Figure 6: Excerpt from a network of genes associated with Crohn Disease. On the right hand side a list of small molecules and drugs can be seen that is associated with the transcription factor NFKB1. Different colourings of genes refer to their expression level derived from an RNA-Seq experiment.
With all variations and in particularly SNP’s and CNV’s, incorporating known biomedical knowledge will allow the move to clinical use of NGS, again for specific disease annotation databases, such as the Genetic Association Database (NIH), GeneAtlas (INSERM) and COSMIC (Sanger) (figure 7).
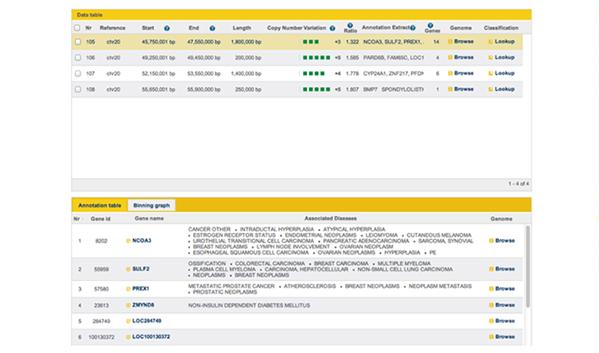 Figure 7: Screenshot from GGA showing disease associations with genes within a CNV region on chromosome 20.
Figure 7: Screenshot from GGA showing disease associations with genes within a CNV region on chromosome 20.
 ,
"...combining all these findings and presenting them in an intuitive visualization environment, researchers and clinicians can interactively relate experimental results to other data ..."
 ,
By combining all these findings and presenting them in an intuitive visualization environment, researchers and clinicians can interactively relate experimental results to other data and to see the result in an understandable format. Deep, extensive, high quality data content as well as innovative visualization tools are therefore critical components of each of these steps and required to enable new biological insights.
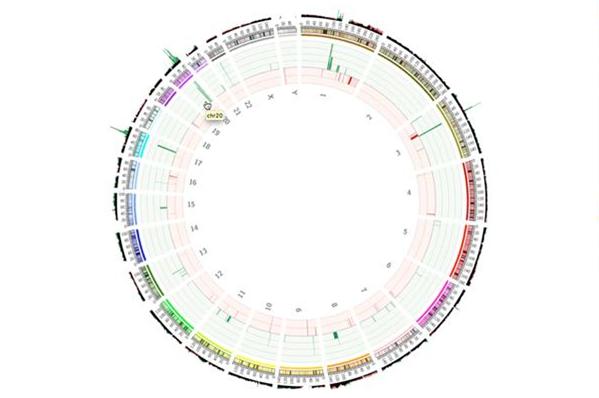 Fig 8: Circos plot showing CNVs across the whole genome.
Fig 8: Circos plot showing CNVs across the whole genome.
Turning results into medical action
With lowered sequencing costs, the increased up take of benchtop sequencers, and the emergence of multiple strategies for data analysis, the medical application of NGS technologies is a natural next step. However, if medical action is to be based on scientific results, several issues need to be addressed:
- Sequencing platforms must provide data that is both reliable and reproducible.
- Analysis pipelines must also be reliable and reproducible. Additionally, they will have to be standardized and certified.
- Medically relevant disease databases need to be integrated with existing data content and their biologically relevant counterparts.
If these and other as yet unforeseen requirements are met, NGS has the potential to become a significant technology in the enablement of personalized medicine.
About the author:
Mark Hughes is currently working for Genomatix as Director of Business for UK and Northern Europe. Having worked in high throughput genomics and particularly analysis of this data since his PhD he has followed this fast advancing field for nearly 20 years.
Prior to starting with Genomatix he worked with some of the leading software providers in the genomics field including Silicon Genetics (now Agilent), GeneGo (combining all these findings and presenting them in an intuitive visualization environment, researchers and clinicians can interactively relate experimental results to other data now Thompson Reuters), Accelrys while also running his own scientific consultancy. As well as extensive work in Europe he has also worked extensively in the US , Asia and Oceania giving him a global view on requirements within genomics.
Email: hughes@genomatix.de
LinkedIn: http://www.linkedin.com/pub/mark-hughes/7/96a/30b
How can NGS be used within personalized medicine?