How GenAI is cracking the toughest targets in drug discovery
There’s no doubt that large language models and generative AI tools have taken the world by storm. Their ability to create, automate, and analyse is continuing to transform the way we work and interact as never before, but the real revolution in generative AI isn't content, it's cures.
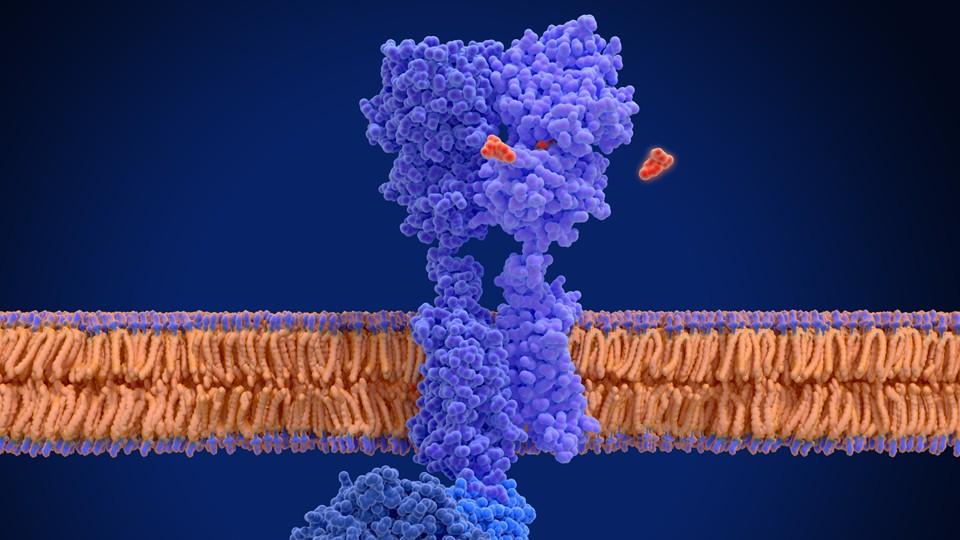
Generative AI has the potential to solve one of the toughest and longest-standing problems of drug discovery and development: designing effective antibodies for G‑protein coupled receptors (GPCRs), a complex and unstable class of proteins that play a vital role in cellular communication, making them prime targets across multiple diseases, including different cancers, Alzheimer’s, cardiovascular diseases, and endocrinology.
Over the years, scientists have struggled to design therapies that can successfully target these receptors – in fact, roughly two thirds of all GPCRs have never been successfully targeted in drug development.
However, this deadlock is now being broken. Thanks to advancements in generative AI, researchers are now able to design lead antibody candidates for previously “undruggable” GPCRs.
By predicting binding interactions and optimising therapeutic properties before lab work even begins, the technology helps unlock new possibilities across drug development pipelines, in an area where over 227 disease-linked GPCRs still lack effective treatments. This innovation could reshape how scientists approach complex targets and, ultimately, accelerate the development of life‑changing therapies for patients with unmet clinical needs.
Traditionally, antibody discovery has required years of experimental trial and error to identify candidates with the right combination of binding, potency, and developability. Generative AI redefines this process by learning the underlying “grammar” of protein sequences and structures, much like a language model learns syntax and semantics. Instead of predicting the next word, these systems can propose novel antibody sequences and structures, optimise binding interfaces to a specific target, and prioritise candidates predicted to have strong affinity, specificity, and favourable development properties.
This unprecedented convergence of computational methods and biology raises important questions. What exactly makes GPCRs such a tough nut to crack? And how can generative AI finally make these undruggable targets, druggable?
GPCRs: Unravelling the enigma of drug design’s shapeshifters
Much like the Greek sea god Proteus, GPCRs are notoriously elusive, with the ability to adopt multiple conformations, thanks to their dynamic structural plasticity.
This forms part of the so-called “iceberg challenge” and the hidden complexities of biological targets, where most of a receptor's structure remains buried deep within the protein or cellular structure, leaving only a minute fraction accessible for drug binding.
In this sense, their shapeshifting quality is both a blessing and a curse; it allows GPCRs to regulate an extraordinary range of physiological processes, making them powerful drug targets. However, it also makes them particularly challenging to study and target with precision.
Despite these obstacles, hope is very much on the horizon. The key to success lies in designing molecules, particularly antibodies, that selectively recognise specific GPCR conformations and epitopes using generative AI technology.
Making the undruggable druggable
The generative AI revolution is speeding up the journey from lab to patient like never before. At the heart of this is a new technology that designs ‘epitope-specific libraries’ using highly detailed structural and sequence data. These libraries consist of millions of highly focused collections of antibody sequences, which can be tailored to bind precise epitopes on target proteins, or designed for specific antibody properties such as affinity, cross reactivity, selectivity, specificity, function, and developability. Enabling the development of highly effective antibodies against targets that are traditionally considered undruggable.
This isn’t a typical trial-and-error drug discovery method; it allows far higher predictive accuracy, significantly boosting the chances of finding antibodies that will form viable therapeutic candidates. This condenses a process that takes up to two years with traditional drug discovery methods of random sampling, down to a few months.
By blending the power of biology and AI algorithms, this opens doors to therapies that were once out of reach, changing the landscape of drug discovery and bringing hope to patients with life-limiting diseases.
Mapping out a new future for drug design
Generative AI is already redefining what’s possible in antibody target development, but its potential spans the entire length of the drug development process. It has the potential to dramatically shorten discovery timelines, reduce failure rates, and predict molecular interactions, such as binding affinity, paving the way for more personalised therapies shaped by individual molecular and genetic profiles.
This acceleration is translating into potential breakthroughs for conditions that have resisted existing treatments, ranging from aggressive cancers to rare genetic disorders, opening new therapeutic avenues across oncology, neurology, and beyond. However, more work is needed to fully realise these goals. The culmination of these efforts remains a distant goal, and true breakthroughs at scale are not yet imminent. Access to more and better-quality data, refined models, and infrastructure and deeper biological insights will pave the way forward to achieving this vision.
Nevertheless, we can look forward to a future where AI doesn’t just design molecules, it promises to rewrite the drug discovery rulebook, empowering researchers to design smarter, faster, and with unprecedented molecular precision. In this brave new world, generative AI doesn’t simply imagine cures; it helps make them real.
About the author

Michael Mahdavi is associate director of business development at Antiverse, where he leads strategic partnerships and works at the intersection of science and business to translate the company’s AI-driven antibody design platform into impactful collaborations and programmes. He holds an MSc in Biochemistry and brings extensive research experience in high-throughput drug discovery.