Study finds hundreds of overlooked cancer drug targets

Researchers from the Wellcome Sanger Institute and partners have carried out a comprehensive analysis of cancer cells, identifying 370 ‘priority’ targets that could be used to develop new drug therapies.
The findings – published in the journal Cancer Cell – came from a systematic look at ‘multi-omic’ information on 930 cell lines taken from 27 different tumour types, including breast, lung, and ovarian cancers, backed up by machine learning technology.
They represent one step closer to the creation of a complete ‘cancer dependency map’ of every vulnerability in every type of cancer, according to the team, which included scientists from public-private target validation initiative Open Targets and partners from other groups, including the Broad Institute in the US.
The new paper covers the second iteration of the dependency map, which was first published in 2019 and provides a “panoramic view of what enables cancer cells to grow and survive,” according to the scientists.
The project uses CRISPR-Cas9 screening to knock out all the genes expressed in the cancer cells, one at a time, to see how they affect their ability to function, hoping to identify those that can be exploited to kill the cells.
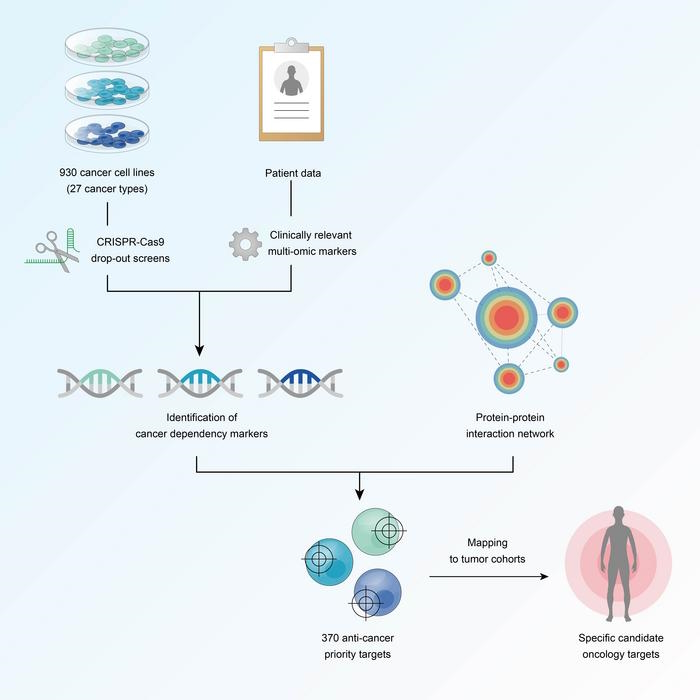
A key objective of the project is to find ‘genetic dependencies’ – the genes, proteins, or cellular processes that cancer cells rely on to survive – that could be harnessed to make new therapies.
Those dependencies are then linked to clinical markers to identify patients for whom those therapies would be most effective, and the dependency-marker pairs are examined to see how they fit in with what is known about molecular interactions in cancer cells.
That can throw out new targets for drug discovery, but also suggest ways in which existing drugs might be re-deployed to treat other forms of cancer – and hopefully help to improve on the current 90% failure rate in new drug development.
Dr Francesco Iorio, co-lead author of the study from the Computational Biology Research Centre of Human Technopole, said the work had created the most comprehensive map of the ‘Achilles heel’ of human cancers.
“We identify a new list of top-priority targets for potential treatments, along with clues about which patients might benefit the most – all made possible through the design and use of innovative computational and machine intelligence methodologies,” he added.
With over 20,000 potential anti-cancer targets in the genome, determining which are suitable to target for specific types of cancers and patients is a significant challenge, and Dr Mathew Garnett, co-lead author at the Wellcome Sanger Institute and Open Targets, said the map “will help drug developers focus their efforts on the highest value targets to bring new medicines to patients more quickly.”
The research was also hailed by Cancer Research UK, which supports the dependency map project.
“Two people might have the same type of cancer, but their diseases can behave differently. That is why we need precision medicine,” commented Dr Marianne Baker, science engagement manager at the charity.
“Giving people treatments for their unique cancer can improve the odds of success and help more people affected by cancer live longer, better lives.”
Photo by Sangharsh Lohakare on Unsplash













