Human ‘pangenome’ could usher in better therapies
It’s been 20 years since the Human Genome Project published its first blueprint of human gene sequences, starting a new era of medical research, but now scientists have gone one better.
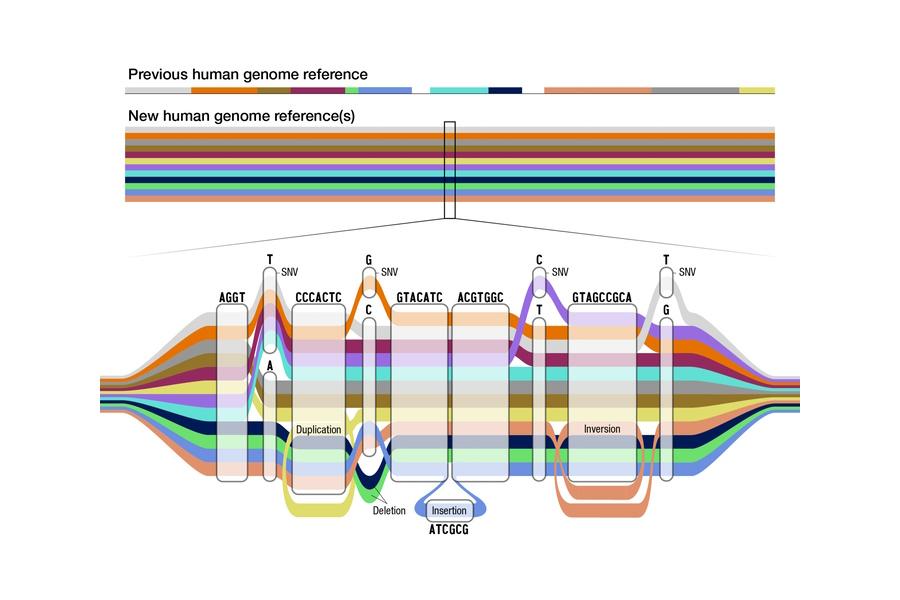
Enter the pangenome, a map that tackles one of the key limitations of the first iteration – namely, its lack of diversity, as most of the sequencing was done on DNA from one American male with European and African ancestry. It also covered around 92% of genetic material, with the remainder uncharted.
The first draft of the pangenome fills in those missing sections of the map, but also covers distinct genomes from 47 individuals from a diverse range of ancestries from all continents in the world, with the exception of Antarctica.
There’s more to come as well, with the researchers behind the effort planning to increase the number to 350 individuals by the middle of next year, and extend the work even further through collaborations with scientific organisations around the world.
The payoff will be the ability to “accelerate clinical research by improving our understanding of the link between genes and disease traits,” according to Yale University scientist Wen-Wei Liao, co-first author of the paper.
That, in turn, could lead to new therapies that will be effective for a wider range of people and diagnostics that will work regardless of an individual's racial or ethnic heritage. The work ties in with other initiatives, for example, trying to ensure that a higher diversity of subjects are recruited into clinical trials.
The pangenome project – published in the journal Nature – relies on the use of advanced computational techniques to align the various genome sequences from the subjects, coming up with a map that represents many different versions of the human genome sequence at the same time, and according to the researchers covers more than 99% of the genome with more than 99% accuracy.
By numbers alone, it adds 119 million base pairs – the building blocks of DNA – to the library of 3.2 billion previously known base pairs that make up the human genome.
Crucially, it will also allow researchers to identify larger genomic variants called structural variants more accurately, thanks in part to advances in long-read DNA sequencing, technology which can read longer stretches of the DNA at a time.
Structural variants are important to molecular biology and medicine, as they play a role in various diseases and regulation of gene expression, amongst many other functions.
Mobin Asri of the University of California Santa Cruz, also co-first author of the paper, said the pangenome will make it possible to “find variants that were not identified using previous methods that depend on linear reference sequences.”
“Basic researchers and clinicians who use genomics need access to a reference sequence that reflects the remarkable diversity of the human population,” said Eric Green, director of the National Human Genome Research Institute (NHGRI) in the US that funded the work.
“This will help make the reference useful for all people, thereby helping to reduce the chances of propagating health disparities,” he added.
“Creating and enhancing a human pangenome reference aligns with NHGRI’s goal of striving for global diversity in all aspects of genomics research, which is crucial to advance genomic knowledge and implement genomic medicine in an equitable way.”












