Epistemic AI launches biomedical GPT at BIO
One of the most compelling things about artificial intelligence chat technologies like ChatGPT and AIChat is their ability to generate human-like responses - but that may not be the best approach when trying to apply them to complex applications like biomedical research.
To make the technology more relevant for researchers, companies have started to develop new platforms with a deeper understanding of the language and processes used in life sciences - notably, Microsoft with its recently-unveiled BioGPT - which aim to be more useful to researchers.
At BIO this week, Boston-based tech company Epistemic AI officially launched its own entry into the embryonic category, EpistemicGPT, with the promise to allow researchers to “interact with highly complex data in a way that was unimaginable until now.”
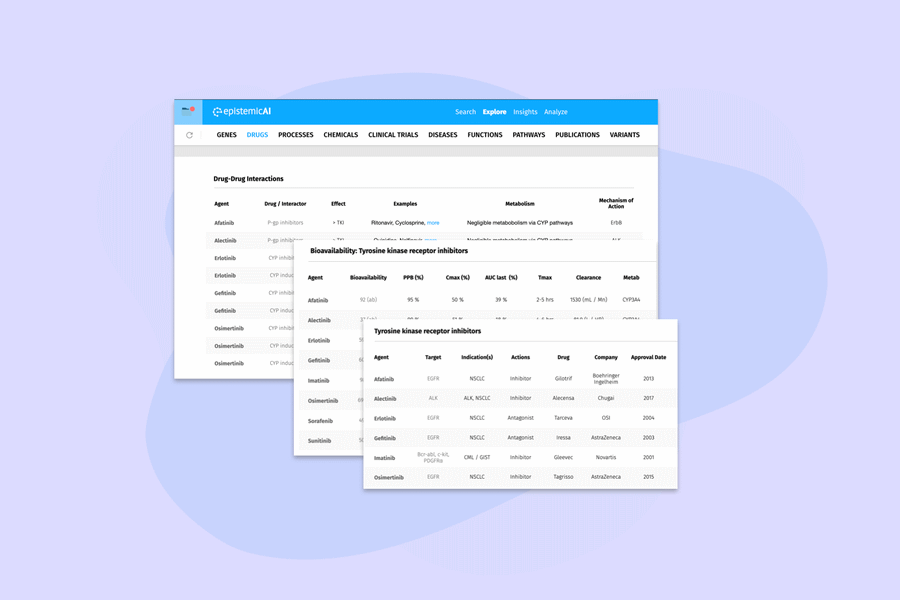
The platform uses large language models (LLMs) and a curated network of billions of biomedical information sources or knowledge graphs (KGs) to help researchers tease out possible connections between different pieces of data.
It can be used to connect drugs and chemicals to targets and indications in drug discovery, help with the design of clinical trial protocols, and sift through ‘omics data to connect gene variants with disease biomarkers, according to Epistemic AI.
Users can also test hypotheses and lead optimisation, prepare documents for trial planning, create market landscape reports, and investigate regulatory approvals “within minutes”.
The platform - which will be made available for free to individual scientists for the time being - is due to be released in beta to a select group in the coming weeks.
“Our goal is to create better tools that can help cure disease,” said Stefano Pacifico, the company’s CEO and co-founder.
“EpistemicGPT provides domain-specific biomedical evidence, with links to original documents and data, removing the concern of unreliable or fabricated information often found with ChatGPT,” he added.
EpistemicGPT has been designed to provide additional functionality, as well. For example, a query about drug resistance cancer results in the creation of a set of workflows that reveal pertinent articles, drugs, and genes.
It is already being used by researchers at Cincinnati Children’s Hospital Medical Center to support their research in cystic fibrosis (CF) and other respiratory diseases and by the Cold Spring Harbor Laboratory (CSHL) as a tool for use by its research staff across multiple disciplines.
Last year, it was cited in an academic research paper study on myocilin, which has been linked to the sight-robbing disease glaucoma, to reveal functional relationships between the glycoprotein and 100 up- and down-regulated genes in animal models.











